1. Introduction
The human race produces about 2.5 quintillion bytes (1018) of data every day, with over 90% of it having been produced over the past five years [1]. Though small in comparison, hospitals create 50 petabytes (250) of data per year, with 97% of it going unused [2]. In this datadriven age, Artificial Intelligence (AI) using deep-learning (DL) has proven and continues to be an effective method for sorting and categorizing data, while recognizing intriguing patterns.
Within medicine, AI has already been applied to the fields of radiology, neurology, orthopedics, pathology, ophthalmology, and gastroenterology [3].Using a database of images, AI has successfully been used to identify cardiovascular disease risk, diabetic retinopathy, and melanoma [4-6]. Additionally, AI has utilized CT scans to determine tumor volume in patients with hepatocellular carcinoma [7]. While AI has also been applied to patient demographics and risk factors, much of the published AI research in gastroenterology has been in the field of medical image analysis (MIA) - a category of AI which analyzes different characteristics of an image to aid in diagnosis, prognosis, and intervention [8].
In the field of gastroenterology, colonoscopy is one of the most routine procedures and is utilized to screen and monitor for colorectal cancer, the second leading cause of death in the United States [9,10]. Each physician performing this procedure is assessed by their adenoma detection rate (ADR), the percentage of colonoscopies performed where at least one adenoma is detected, with an ADR goal > 25% [11]. This metric is important because it has been shown that a 1% increase in ADR is associated with a 3% decrease in CRC [12]. However, some research has shown that up to 27% of polyps are missed [13,14].
Given the importance of ADR and the amount of video and imaging colonoscopies produce, research in AI application to polyp detection and characterization has been very popular. In the past twenty years, AI along with computer-aided detection and diagnosis (CADe and CADx) techniques have been optimized with following three goals for polyp detection 1) high sensitivity 2) low false positive rate 3) low latency so polyp detection can be done in real time [15]. These methods not only aim to increase ADR, but also identify hyperplastic vs. adenomatous polyps.
As our technology progresses and AI becomes more advanced, it is inevitable that AI will play a large role in colonoscopies. Thus, in this review article, we will discuss the basics of AI, how it has been used to detect and characterize polyps, and the future direction and complications of it.
2. What is Artificial Intelligence (AI)?
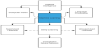
In 1950, Alan Turing described how computers were as intellectually capable as humans, and by 1956 John McCarthy introduced the word Artificial Intelligence (AI) [16,17]. AI can be defined as a machine demonstrating human intelligence in terms of cognitive functioning, learning, and problem solving [18]. These machines, which were first comprised of “if, then rules,” have now become an aggregation of interconnected intricate algorithms that function like a human brain [19]. The way these algorithms are designed can be broken down into subsets within AI, the most common being machine learning (ML) (Figure 1) [20]. Common terminology associated with Artificial intelligence can be found in (Table 1).
ML automatically creates mathematical algorithms to predict future outcomes and patterns from inputted data which can then be applied to future scenarios without human intervention [20]. Within ML, there are two types of learning: unsupervised learning and supervised learning. In unsupervised learning, unlabeled data is fed into the computer and commonalities are found within the dataset. For example, this has been used in predicting glycemic responses to foods and in personalized medicine through analysis of patient history, labs, and imaging [22,23]. In supervised learning, labeled data is used to create algorithms to label future data sets through clustering and grouping. This type of learning has been used to predict patient outcomes [24,25].
Since its invention in 1952, ML has evolved into what is now known as deep learning (DL) - a composition of algorithms that make up an artificial neural network (ANN). DL transcended ML into practical use because of its multilayer back-propagation algorithm, which allows it to cross reference parameters in each layer and create outputs more efficiently (Figure 2) [21]. In essence, smaller patterns and details can be identified through DL layers with earlier layers having generic details and later ones being more specific [26].
In the polyp detection, DL has been utilized with different Convolutional Neural Networks (CNN) to create computer vision, a mechanism to process a series of images or videos (Figure 2). CNNs work by extracting multiple specialized features of an image to create multiple maps, which are then aggregated together to produce an output [20]. This can be compared to the way the human brain processes an image. Basic parts of an image like boundaries and light and dark areas are first recognized. These basic parts are then combined to make simple shapes and eventually complex ones in subsequent layers. These “convolutional” layers dissect an image into easily processed parts, which later feed into a “pooling” layer to conserve data size and reduce noise. Ultimately, this process creates an algorithm which identifies the probability a future image will contain similar features without the programmer describing the specific feature. This is important in polyp detection because endoscopists tend to have difficulties in describing the polyps in detail [27].
3. Colorectal Polyp Detection
The rate of polyps missed on colonoscopy is as high as 25% [28]. This may be due to poor bowel prep, appearance of the polyp, or the endoscopist’s inspection technique. Improvement in polyp detection may lead to an overall decrease in colon cancer.
By the early 2000s, researchers had begun investigating the applications of ML in polyp detection. Using recorded videos and images, hand-crafted algorithms targeting polyp features such as color, shape, or texture were initially used for many of the ML algorithms. Karkanis et al. developed one of the first polyp detection softwares called CoLD (Colorectal Lesions Detector) using texture and color wavelet covariance to differentiate normal from abnormal tissue [29,30]. A few years later, this group’s algorithm identified adenomatous and hyperplastic polyps with 93.6% sensitivity and 99.3% specificity [31,32]. Similarly, Tjoa and Krishnan used a texture spectrum and color histogram, while Zheng et al. utilized texture and luminal contour to identify abnormal tissue [33,34]. While these methods produced results with high sensitivity and specificity, they were limited by their slow processing times and their inability to detect atypical polyps or differentiate false positive non-polyp lesions like stool.
As DL became more developed in the field, larger data sets were being analyzed to produce algorithms that could be applied to other videos and pictures to test for polyp detection. Some notable ones being Wang et al. applying DL polyp detection software to 5545 retrospective images that had been previously diagnosed by endoscopists and 27461 prospective colonoscopy images from 1235 patients [35]. Additionally, Misawa et al. used 105 polyp positive videos and 306 polyp negative videos to develop an algorithm with 94% accuracy, but with a false positive of 60% [36].
Application of CNN in polyp detection also proved promising. Billah et al. developed an algorithm with 99% sensitivity and specificity on a public dataset by using a CNN and their own algorithm [37]. However, processing time was not mentioned in their study. Likewise, Zhang et al. utilized millions of images from ImageNet, a database of everyday objects like fruits and cars, to develop an algorithm which achieve a polyp detection sensitivity of 98% [38]. When comparing this CNN to an endoscopist, it outperformed the endoscopist 86% to 74% in terms of accuracy. Additionally, Li et al. used a CNN on 32305 colonoscopy images and developed an accuracy of 86% and sensitivity of 73% [39]. All of these studies show promising results on how DL and CNN can be used to identify polyps.
The most important application of these AI algorithms is in realtime detection. The goals in real time detection are high sensitivity, high specificity, with low latency between detecting the polyp and when it appears on the secondary screen. Typically, colonoscopy videos run at 25-30 frames per second or 33-40 ms per frame. With traditional ML, Tajbakhsh et al. was able to base their algorithm off of hybrid context-shape, where context was used to filter out non-polyps and shapes were used to identify polyps [40]. This approach yielded 88% sensitivity with a latency of 0.3 seconds. However, these results are limited because they were done retrospectively on 25 polyps. Likewise, Fernandez-Esparrach et al. used energy maps from the localization of polyps and their boundaries and achieved a sensitivity of 70.4% and specificity of 72.4% in 24 videos containing 31 polyps [41]. Finally, Wang et al. utilized “polyp edges” to achieve a detection rate of 97.7% with 36 false positives coming from folds, the ileocecal valve, appendiceal orifice, and areas of colon with residual fluid [42].
One of the most promising applications of CNN in real-time detection is from Urban et al. where ImageNet was used to develop the algorithm and then subsequently applied to multiple colonoscopy images and 11 videos [43]. Their method achieved 97% sensitivity, 95% specificity, 96% accuracy, with the algorithm executing at 10ms per frame. On top of that, the CNN method detected 17/45 polyps that were missed by the endoscopists. Byrne et al. also developed a deep CNN for real-time detection using narrow-band image video frames and videos from colonoscopies to achieve a sensitivity of 98%, specificity of 83%, and accuracy of 94% on 125 videos of 106 polyps [44]. Interestingly, Yu et al. developed a three-dimensional CNN using spatiotemporal features of colonoscopy videos and was able to reduce the number of false positives from a previous 2015 data set [45]. These advances in CNN related research in regard to high sensitivity, specificity, low lag time, and different methods of polyp detection show promise to application in real time detection.
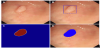
To date, only three real-time in vivo studies have been performed. Klare et al. utilized software which analyzed color, texture, and structure with 50 ms lag time on 55 routine colonoscopies [46]. Their results showed comparable results between the endoscopist and the software. The endoscopists polyp and adenoma detection rates were 56.4% and 30.9%, while the software’s detection rates were 50.9% and 29.1%, respectively. The software also did not detect any additional polyps that the endoscopist missed. Wang et al.’s in vivo study also produced similar results [47]. In a randomized controlled trial of 1058 patients with their CNN, their software had had an ADR of 29.1% vs 20.3% of the endoscopists (Figure 3). This difference was mainly due to the polyps being smaller than 10 mm and a higher portion of them being hyperplastic polyps. Additionally, only there were only 39 false positives (0.075) in the trial, which is on average in a regular colonoscopy. Finally, Repici et al. performed a randomized trial with 685 people of AI for polyp detection in real-time colonoscopy for indications of screening, surveillance, or fecal immunochemical test positivity [48]. Their system improved ADR from 40.4% to 54.8% when compared to a control, especially in those <5 mm in size and those with a diameter 5-9 mm. Most importantly, their system did not increase withdrawal time.
4. Colorectal Polyp Characterization
When polyps are located, they must be characterized to determine if they must be biopsied. In the field of AI, this is referred to as computeraided diagnosis (CADx). Typically, when polyps are identified, they can be left alone, resected and discarded or sent to the lab for histopathological examination. These strategies known as “diagnose and disregard” and “resect and discard” have been suggested to save millions of dollars every year in the U.S [49,50]. Current techniques being used to characterize polyps include magnifying narrow band imaging (NBI), endocytoscopy, laser-induced fluorescence spectroscopy, and Auto-Florescent endoscopy (Table 2) [51]. However, amongst these techniques, it has been shown that there is much variability amongst users [52-54]. The use of CADx may decrease this discrepancy between endoscopists.
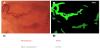
NBI is an optical modality that uses blue and green light to look at polyp vessel size and pattern (Figure 4). Using the color, vessels, and surface patterns, endoscopists can differentiate between hyperplastic and adenomatous polyps [55]. In 2010, Tischendort et al. used Narrow Band Images (NBI) images to create a classification model based off of a colorectal polyp’s vascular pattern with 91.9% accuracy [54]. Then in 2011, Gross et al used NBI images to differentiate polyps < 10 mm with 95% sensitivity, 90.3% specificity, and 93.1% accuracy [56]. Initial studies done by Takemura et al. with still images was able to achieve a sensitivity, specificity, and accuracy of 98%, 98%, and 98% for neoplastic lesions, respectively [57]. A later study with 41 patients and 118 colorectal lesions by the same group were done in real time, which yielded a sensitivity, specificity, and accuracy of 93%, 93%, and 93.2% [58]. While the sample size was relatively small, this proved important because it met the criterion for the “diagnosis and leave” strategy, as well showing the effectiveness of CADx in real-time compared to human diagnostics.
Endocytoscopy (EC) is another form of optical diagnosis that has benefited from CADx development. In EC, endoscopists look at the microvascular structure and surface epithelial at the cellular level [59]. This method of optical diagnosis has been of particular interest because it provides focused and fixed images for AI analysis. The earliest study involving EC comes from Mori et al. in 2015 when they used images from 176 polyps and 152 images [60]. Their CAD system produced a sensitivity and specificity of 92% and 79.5% vs. 92.7% and 91% when compared to endoscopists. This same group most recently performed a prospective study with 466 polyps from 791 patient, which achieved a sensitivity and specificity of 97% and 67% and a negative predictive value of 93.7% [61]. This study, however, elucidated the difficulty of identifying diminutive non-neoplastic polyps, as they were only correctly identified 70% of the time. Widespread use of EC is limited by the availability of EC scopes.
In laser-induced fluorescence spectroscopy, polyps are determined to be non-neoplastic or neoplastic by the light it reflects back once it shown laser light. Kuiper et al. prospective research showed 83% sensitivity, 59.7% specificity, 71% positive predictive value (PPV), and 74% negative predictive value (NPV) [62]. However, Rath et al. also investigated the same method and produced a NPV of 96.1% [63]. Though the results varied differently, future studies in this field appear promising as this technology would allow endoscopist to remove polyps immediately after pathology prediction without having to exchange instruments.
Auto fluorescence imaging (AFI) is another strong candidate for AI implementation. In this diagnostic modality, the endogenous fluorophores of the mucosa are elicited through light, from which a green/red image is produced and analyzed by the computer. Aihara et al. investigated the use of AI in AFI imaging on 32 patients with 102 polyps and showed 94.2% sensitivity and 88.9% specificity [33]. Additionally, Horiuchi et al. conducted a prospective trial of 429 polyps in 95 patients and produced results of 80% sensitivity, 95.3% specificity, 85% PPV and 93.4% NPV [64].
Real-time analysis done with white light without advanced imaging may also be possible with the help of AI. Byrne et al. developed a deep CNN on white-light and NBI colonoscopy videos [65]. This algorithm was then tested on 125 videos and achieved 94% accuracy in classification in 106 of the videos. Of the 106 videos, the CNN was able to detect adenomas with 98% sensitivity and 83% specificity.
5. Future Direction and Complications
Despite the great advancements in polyp detection and characterization, the field has been slow to adapt the technology. One of the reasons is that much of the research has been done retrospectively and at single centers with a select data set. This may be a problem in the future as this could lead to over fitting - when a model creates an algorithm that cannot be generalized to other data sets. Additionally, having a limited data set may also lead to spectrum bias - when the data model set does not accurately represent the patients it will be used on in the future. These two problems are important to note as most of the recent studies have taken place in China and Japan, where colorectal cancer rates are much lower compared to the U.S.
While DL attempts to mitigate these problems, it does not guarantee to fix it. One way to overcome this problem is to expand the available data set and collaborate with other centers. Currently, CNNs are being pre-trained with the ImageNet database, which has over 14 million annotated images, before being applied to polyp detection [66]. When looking at most of the studies on polyps, these studies only use hundreds to thousands of annotated polyp images. In fact, the largest data base of annotated polyp images contains less than 20,000 images [67]. Pooling image data sets between centers may prove to be beneficial for more accurate development of CNNs.
Ethical and liability concerns about AI must also be addressed before mass adoption. For example, if an endoscopists misdiagnoses a polyp, who is liable - the endoscopist or the manufacturer of the device? Are there any inherent demographic biases that the machine is producing which cannot be detected by physicians and programmers? The “black-box” nature (lack of interpretability and explainability) of most of these machines make these questions more difficult to answer and may make physicians hesitant about the technology. Arguments that physicians will rely too heavily on AI or that AI will replace physicians have also been brought up for discussion.
With the increasing number of physician burnout, musculoskeletal injury from endoscopy, and shortage of human resources, AI may help overcome many of the negative factors that affect diagnostic screening by improving lesion detection, reducing medical error, and improving efficacy. Additionally, with the growing demand for precision medicine from patients and physicians, AI may help pave the way. Although much of the research being done aims for high sensitivity and specificity for polyp detection and characterization, high sensitivity may only be needed for improved detection.
Additional research must be done on live colonoscopies and also into physician satisfaction, cost effectiveness, and clinical outcomes related to the mass adoption of the technology. Some outcomes to be studied include adenoma detection rate and withdrawal times. Additionally, gastroenterologists must pay attention to AI application in other fields of medicine and observe the pitfalls and successes. Finally, there must be a clear path to FDA approval for future device acceptance.
6. Conclusion
The field of gastroenterology has made incredible leaps in terms of integrating AI into the field. In particular, developments of AI in polyp detection and classification have shown promising results. The strengths of AI include the ability to process and analyze large amounts of data much faster than humans, though some barriers to more research is limited by the amount of publicly available annotated data for AI processing. Current directions of the field include development of more sophisticated CNNs and real-time prospective and randomized controlled trials in vivo studies. These steps will not only optimize AI techniques, but also continue to show the utility in the clinical setting. Although computers may be more accurate and precise in diagnosis and detection of disease, physicians will still be needed to synthesize the information and communicate it with the patient. For all these reasons, AI promises to accompany physicians in providing better care to patients.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgments
This work is supported in part by Dr. Moro O. Salifu’s efforts through NIH Grant # S21MD012474.