1. Introduction
Forcipomyia taiwana is a tiny (1.0-1.5 mm) bloodsucking midge that is widely distributed in both urban and rural environments across Taiwan and parts of Southern China [1]. Female F. taiwana are known to feed during the day from the exposed body surfaces of humans, and their bites can cause intense itching, swelling, skin lesions, and even serious anaphylactic reactions; overall, about 60% of individuals bitten by F. taiwana will develop varying degrees of hypersensitivity [2]. Although no midge-borne diseases have been reported in the Taiwanese population as yet, F. taiwana bites can be an irritating nuisance at best, or can result in anaphylaxis or cellulitis at worst. Therefore, an understanding of the factors that enhance or inhibit F. taiwana feeding behavior may be useful to controlling the current and future impact posed by these biting midges.
Recent research has increasingly shown that the feeding behavior of bloodsucking insects is not purely affected by interactions between the insects and human hosts alone; for example, the Plasmodium malaria parasite has been reported to influence the feeding behavior of female Anopheles gambiae mosquitoes [3,4], and studies show that human skin microbiota can affect the feeding preferences of mosquitoes [3-6], in part by altering body order [3]. However, it is as yet unclear whether F. taiwana demonstrates odor-mediated host-seeking and feeding behavior. More importantly, a suitable animal model to examine F. taiwana feeding patterns has not yet been developed. Mice (Mus musculus) have been used as animal models to examine feeding behavior in mosquitoes (Culexpipiens) [7], or to assess the effectiveness of repellants against laboratory-cultured mosquitoes [8]. Unfortunately, due to the differences in proboscis structure between mosquitoes and midges, mice cannot be used as animal models to examine midge feeding behavior, as midges lack the elongated proboscis of mosquitoes and need to be in contact with exposed skin in order to use their mandibles to draw blood. A recent publication presenting the development of a murine model for F. taiwana allergies utilized intradermal application of midge extract, with no actual biting behavior involved [9]. Previously, in order to elucidate the midge life cycle from eggs to pupae, researchers described the use of their own blood to nourish female midges; however, although midge-insensitive humans may be relatively unaffected by repetitive feeding, it has been reported that long-term feeding may induce hypersensitivity to biting midges in humans [1].
In this study, we describe the use of New Zealand White rabbits as an animal model to investigate F. taiwana feeding behavior. New Zealand White rabbits are widely used in animal experiments and can be consistently managed in the lab. In addition, the ears of New Zealand White rabbits stand straight up and only have a short fur coating, conditions that are much more conducive to F. taiwana feeding as compared to other laboratory animals. We collected human sweat from two different individuals, one of which exhibited preferred feeding by F. taiwana. Sweat from each individual was respectively applied to the right ears of two New Zealand White rabbits, with the left ears serving as controls. We found that F. taiwana displayed a distinct feeding preference for rabbit ears to which human sweat had been applied; moreover, sweat from individuals with less diverse skin microbiota appeared to be more favored by female biting midges. The results indicate that skin microbiota may also influence the feeding behavior of F. taiwana, and our findings could have interesting implications for the development of biorepellants against this biting midge.
2. Methods
2.1 Ethics Statement
Human subjects were enlisted for the purpose of sweat collection only, and were managed in accordance with the Declaration of Helsinki. Prior informed consent was granted by all human subjects for participation in sweat collection for the purposes of this study. The animal experiments in this study were designed and performed in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals, 8th Edition (https://grants.nih.gov/grants/olaw/Guide-for-the-Care-and-use-of-laboratory-animals.pdf) and the Guide for the Care and Use of Laboratory Animals of Tzu Chi University, Hualien, Taiwan (http://www.lac.tcu.edu.tw/?page_id=474). The protocol was approved by the Institutional Animal Care and Use Committee of Tzu Chi University (Permit Number: 104042). Two New Zealand White rabbits of similar age were bred in the Tzu Chi University Laboratory Animal Center, and were housed in a specific pathogen-free facility with individual cage ventilation systems. Study rabbits had free access to autoclaved water and food, and were maintained at room temperature (~25°C) on a 12- hour light-dark cycle. Rabbits were allowed to acclimatize for at least one week prior to F. taiwana feeding behavior experiments, and all efforts were made to minimize unnecessary suffering.
2.2 Collection of F. taiwana
Female F. taiwana were collected by the human bait method, as previously described [10]. At least 20 female midges were kept in a 1.5 cm (diameter) × 4.5 cm (height) glass vial with a volume of 10 ml, and the vial was covered with tulle to prevent midges from escaping, but which allowed midges to feed on the ears of study rabbits. A total of five such vials were prepared prior to experiments on midge feeding behavior.
2.3 Human sweat collection and midge feeding experiment
At least 1 ml of sweat was respectively collected from each human subject (A and B) and kept in similar small vials prior to use. Human subject A exhibited preferred feeding by F. taiwana, while human subject B did not have such a profile. For midge feeding experiments, 100 μl of human sweat from human subjects A and B were respectively applied to the right ears of study rabbits A and B, while 100 μl of ddH2O were applied to the left ears. Vials containing female F. taiwana midges were then placed over rabbit ears for 30 minutes, and feeding behavior was observed and recorded. Midge-containing vials were then placed at 4°C for 20 minutes to rest midges for easy observation, after which feeding outcomes were observed under a microscope. The results were then compared with the real-time observation results.
2.4 Total DNA isolation from sweat
The remaining sweat was transferred from storage vials to microcentrifuge tubes, and centrifuged at 10,000 xg for 10 minutes. Total DNA was isolated according to a previously described method, with minor modifications [11]. Precipitates with bacteria and human skin debris were resuspended in 500 μl of Solution I (50 mM glucose, 25 mM Tris-Cl (pH 8.0), 10 mM EDTA (pH 8.0)) with lysozymes (1 mg/ml), and kept at 37°C for 30 minutes. Afterwards, 100 μl of 10% SDS and 20 μl protease K (20 mg/ml) were added, and the mixture was incubated at 55 °C for one hour. An equal volume (620 μl) of phenol/chloroform was then added to extract the DNA, which was subsequently precipitated with 0.1 volume of 5 N NaCl and 2 volumes of ice-cold 100% ethanol at -80 °C for 20 minutes.
2.5 PCR amplification of 16S rDNA
PCR reactions were performed to amplify 16S rDNA, using the following primer pairs as previously described: [12] (i) Bakt341F: 5’-CCTACGGGNGGCWGCAG-3’ and Bakt805R: 5’-GACTACHVGGGTATCTAATCC-3’; and (ii) Bakt-341Fad: 5’-TC GTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNG GCWGCAG-3’ and Bakt-805Rad:
5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGAC TACHVGGGTATCTAATCC-3’. Reactions were performed in 50-μl volumes containing 250 μM dNTP, 0.5 μM of each primer, 1.25 U Taq DNA polymerase, 1.25 U Pfu DNA polymerase, and 1× reaction buffer. PCR conditions were as follows: initial denaturation at 94°C for 5 minutes, followed by 20 cycles of denaturation (94°C for 30 seconds), annealing (55°C, 30 seconds), extension (72°C, 30 seconds), and a final extension step at 72°C for 5 minutes. PCR products were analyzed by electrophoresis, and purified using a PCR clean up kit (Geneaid, Taipei, Taiwan), after which 10 μl of purified products were used as templates to perform another 5 PCR cycles using the second pair of primers, under the same conditions as that used with the first pair of primers. The quality and quantity of extracted DNA were analyzed by agarose gel electrophoresis, as well as by spectrophotometry using a NanoDrop 2000C spectrophotometer (Thermo Scientific, Waltham, MA).
2.6 Metagenomic analysis of microbial diversity in sweat
A total of 206,191 sequences of 16S rDNA were analyzed by mothur v1.34.1 software, using the MiSeq standard operational procedure (http://www.mothur.org/wiki/MiSeq_SOP) for defining OTUs (operational taxonomy units). Sequences shorter than 480 bp were removed, and sequences were grouped into OTUs by using the nonredundant (NR) SILVA v119 database with a minimum sequence identity of 97%. Chimeras were examined and removed by using the command: chimera.uchime. Sequences originating from chloroplasts and mitochondria were removed. Finally, 55,671 sequences from the sweat of human subject A and 47,860 sequences derived from the sweat of human subject B were classified and assigned into OTUs based on SILVA taxonomy. Two communities were randomly subsampled into 47,860 sequences for subsequent community comparison.The representative sequence of each OTU was further classified using the Ribosomal Database Project (RDP) classifier and GenBank database[13]. Microbial diversity was analyzed by PAST v2.17 softwareto determine all diversity indexes.
3. Results
3.1 New Zealand White rabbits as an animal model for F. taiwana feeding behavior
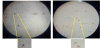
The suitability of New Zealand White rabbits as an animal model to investigate F. taiwana feeding behavior was assessed, with the relatively exposed ears considered to be the most appropriate region for midges to feed upon. Vials containing at least 20 midges were used to perform midge feeding tests. The vials were covered with tulle to prevent midges from escaping but still allow feeding to proceed, and these vials were used to cover the ears of study rabbits for 30 minutes. Vials were then placed at 4°C to immobilize midges, after which midges were examined under a dissecting microscope to assessfeeding outcomes. Results showed that midges with bloated abdomens engorged with blood were widely observed in the treatment groups (Figure 1), but several midges with flat abdomens and no evidence of feeding were observed in the control groups.
3.2 Human sweat induces differential feeding behavior in F. taiwana midges
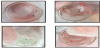
Study rabbits A and B, the right ears of which were respectively smeared with sweat from human subjects A and B, were used to conduct feeding experiments in the Laboratory Animal Center of Tzu Chi University. Human subject A exhibited preferred feeding by F. taiwana, and during the feeding experiment, more midges were observed on the tulle covering of the vial on the right ear of study rabbit A (Figure 2a), which was smeared with sweat from human subject A; however, in other vials, midges were equally distributed around the space (Figure 2b-d).
After the feeding experiment was conducted for 30 minutes, bite marks were found on both right ears of study rabbits. However, bite marks were larger and more visible on the right ear of study rabbit A as compared to study rabbit B, while no significant bite marks were observed on the left ears of both study rabbits (Figure 3). All bite marks subsequently disappeared within one hour after the feeding experiment was concluded.
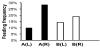
After the feeding experiment, F. taiwana midges were observed under a dissecting microscope to assess feeding outcomes. Bloated midge abdomens engorged with blood were counted to determine the biting frequency for each vial. The vial on the right ear of study rabbit A had the highest biting frequency observed (Figure 4a, (R)), while the vial on the right ear of study rabbit B (Figure 4b, (R)) did not differ greatly from controls (Figure 4a, (L) and 4b, (L)). Considering that the right ear of study rabbit B was smeared with sweat from human subject B, who did not exhibit any preferential feeding by F. taiwana, the results suggest that human sweat can induce differential feeding behavior in such biting midges.
3.3 High skin microbiota diversity attenuates F. taiwana feeding
To understand the differences in F. taiwana feeding behavior induced by the sweat from human subjects A and B, we sought to examine the bacterial populations present in sweat, as this reflects upon the diversity of skin microbiota for these individuals. Total DNA in the sweat from human subjects were isolated and amplified with 16S rDNA specific primers for subsequent next-generation sequencing (NGS) analysis. After data filtering processes, a total of 95,720 valid reads and 279 OTUs were obtained from two groups of samples. Altogether, 189 and 210 OTUs were respectively obtained from the sweat samples derived from human subject A (midge-feeding preference) and subject B (non-preference). These sequences were assigned to 19 different phyla (Figure 5a) and 171 different genera (Figure 5b). The Shannon diversity index, a biodiversity measure that incorporates both richness and evenness, was also calculated for the sweat samples. The Shannon diversity index for human subject B (1.866) was higher than that for human subject A (1.077), suggesting that F. taiwana midges may be attracted to humans with lower skin microbiota diversity.
Bacteria of the Firmicutes phyla predominated in the sweat sample from human subject A, followed by Proteobacteria and Actinobacteria; however, bacteria from these three phyla were distributed equally in the sweat sample from human subject B (Figure 5a). Moreover, Staphylococcus made up 80% of the genera in the sweat sample from subject A, but represent less than 50% of the genera in the sample from subject B (Figure 5b), suggesting that the abundance of Staphylococcus spp. may correlate with the relative attractiveness of human subject A to F. taiwana. In addition, Serratia and Propionibacteria respectively make up 19.6% and 11.5% of the skin microbiota of subject B (Figure 5b), suggesting that these two genera may exert a repelling effect. By contrast, Streptococcus, Neisseria, Derxia, Capnocytophaga and Rothia were present in the sweat of subject A at levels 10-fold higher than subject B, suggesting that these genera may have an attractant effect for F. taiwana.
4. Discussion
There are currently no reports that F. taiwana acts as a disease vector, but their bites do pose a risk of hypersensitivity and anaphylaxis [10]. A murine model of F. taiwana bite allergy has recently been established, using a two-step sensitization protocol that involved the intraperitoneal injection of mice with F. taiwana whole body extract to study the immunopathologic features of midge allergy [9]. However, extracts cannot fully replicate the feeding behavior of F. taiwana, and thus a suitable animal model is needed to investigate the life cycle and potential disease transmission capability of F. taiwana in more detail. In this study, we describe just such an animal model, using New Zealand White rabbits. This model offers researchers an opportunity to examine the natural habits and behavior of F. taiwana under controlled conditions, and without having to rely on human sources for feeding.
It is currently known that the host-seeking behavior of mosquitoes is affected by diverse factors, including CO2 exhalation, host body odor, human leukocyte antigen (HLA) profiling or even beer consumption[14]. Carbon dioxide is a major constituent of exhaled breath, and appears to be an attractant for some mosquitoes; however, carbon dioxide is not a major attractant for the Afrotropical female malaria mosquito, Anopheles gambiae [15], which prefers to exploit olfactory signals emanating from human hosts[16]. Malaria mosquitoes have been reported to be attracted to compounds such as lactic acid, 2-methylbutanoic acid, tetradecanoic acid, and octanal in human body odor[17], and recent studies have assayed 300-400 compounds in human odor with gas chromatography (GC)/mass spectrometry (MS) to identify possible attractants for the Aedes aegypti mosquito[18]; however, further bioassays will be needed to derive definitive conclusions. Nevertheless, it is known that the volatiles that make up human odor are mostly derived from the secretions of skin glands, which are originally odorless to the human nose, but are subsequently converted into odorous and volatile compounds through the enzymatic action of skin microbiota. The body odor of individual humans has been correlated with the presence of specific microorganisms [19]; for example, culture-based studies have shown that human skin microbiota is dominated by corynebacteria, staphylococci, or propionibacteria, of which corynebacteria have been associated with the production of odorous steroids and thioalcohols, while staphylococci are known to metabolize branched aliphatic amino acid to short-chain (C2-C5) volatile fatty acids (VFAs) [6]. In this study, we show that midges are attracted to sweat from an individual with a skin microbiota dominated by staphylococci (~80%), and this raises the possibility that VFAs may be an important attractant for F. taiwana; however, further GC/MS analysis of volatile compounds in sweat and an expansion of the current pilot study to include more individuals would be needed to confirm this.
In summary, we showed that New Zealand White rabbits may serve as an animal model to examine feeding behavior for F. taiwana, and also presented preliminary results from a pilot study that demonstrated human sweat may influence F. taiwana feeding preferences. Our findings suggest that skin microbiota diversity and the presence of specific microorganisms may also play a role in making an individual more or less susceptible to F. taiwana biting, and may have implications for the development of effective biorepellants against biting midges in future.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgments
The authors would like to thank Dr. Hsien-Ming Lee from the Institute of Medical Biotechnology, Central Taiwan University of Science and Technology, and Dr. Ming-Hseng Wang from the Laboratory Animal Center of Tzu Chi University for technical advice. The authors also would like to thank Miss Yu-Hsuan Chiu, Meng-Yun Wen and Mr. Kun-Sheng Hsieh for helping to conduct midge feeding experiments.