1. Introduction
Chlamydia trachomatis is the leading cause of sexually transmitted infections (STI) worldwide. We have analyzed reference and clinical samples that has not previously been represented in C. trachomatis typing schemes [1,2], to expand our knowledge of the global diversification of strain types (serovars) [3-6]. The samples resolved into unique serovars, compared to the gold standard single locus ompA (or omp1) genotyping. Phylogenetic analyses revealed distinct branches for the phenotypic diseases of lymphogranuloma venereum (LGV), urethritis and cervicitis, and a sub-branch for ocular trachoma [7].
Fifteen serovars and variants of C. trachomatis were originally classified by the micro-immunofluorescence (MIF) test which principally distinguished epitopes on the major outer-membrane protein (MOMP) [8]. C. trachomatis exists as classic 15 serovars such as A, B, Ba, C, D, E, F, G, H, I, J, K, L1, L2 and L3. Serotyping correlates well with the four variable domains (VDs of MOMP. The 15 classic serovars can be divided originally into two pathotypes. The ocular pathotypes including serovars A-C are associated with so-called classic trachoma which is still hyper-endemic in Africa, Asia, and South America. Trachoma is still the second major cause of blindness all over the world [9]. The sequences of MOMP gene including four VDs have been determined for all classic 15 serovars [10].
Ocular pathotypes are usually not isolated from the genital tract. Under the genital pathotypes, which causes STI, serovars D-K affect mainly the mucous membrane. It should be pointed out that in the literature, serovars D-K, together with serovars A-C, have been grouped into the so-called trachoma biovar. However, these two serovar groups affect the eye quite differently. Whereas the ocular serovars (A-C) cause chronic ocular infection, which may lead to blindness, the genital serovars (D-K) cause only transient, selflimiting infection in the conjunctival mucous membrane without severe complications.
The PCR assay was used to amplify a large part of the MOMP gene (ompA or omp1) including four VDs and then cataloged restriction fragment length polymorphism (RFLP) was used to identify the serovars from genotypes of C. trachomatis from nasopharyngeal and conjunctival swabs obtained from Japanese infants and neonates with pneumonia and conjunctivitis as reported previously [1,8]. In this study I detected the presence of unclassified serovars of C. trachomatis, by means of PCR-RFLP analysis and sequencing of amplified DNA, obtained from Japanese infants.
2. Materials and Methods
2.1 C. trachomatis reference and clinical samples
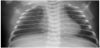
Nasopharyngeal specimens were collected from 15 neonates and infants with pneumonia from 1 month to 1 year old, during the period May 1990 to September 2013. Conjunctival swabs were also collected from 12 neonates under 1 month old with inclusion conjunctivitis during the same period. Diagnosis of pneumonia was based on clinical findings and radiologic confirmation (Figure 1 and Figure 2). Clinical samples were analyzed including the 23 reference strains: A/ SA-1, A/HAR-13, B/TW-5/OT, Ba/AP-2/OT, B/ TW-5/OT, B/Jali-20/ OT, C/TW-3/OT, D/ UW-3/CX, D/IC-Cal-8,Da/TW-448, E/Bour, F/ IC-Cal3, G/UW-57/CX, H/UW-4/CX, I/UW-12/UR, Ia/UW-202, J/ UW-36/CX, Ja/UW-92, K/UW-36/CX, L1/440, L2/434, L2a/UW-396, L3/404.
2.2 RFLP pattern analysis of the amplified MOMP gene for genotyping
The PCR assay to amplify ompA gene and RFLP analysis were used to detect and distinguish serotypes from genotypes of C. trachomatis as reported previously [1,2]. Briefly, an approximately 1 kb fragment of the ompA gene was amplified using primers SERO1A (5'-ATG AAA AAA CTC TTG AAA TCG G-3') and SERO2A, (5'-TTT CTA GAT CTT CAT TCT TGT T-3'). The reaction was performed in a final volume of 90 μl containing 1.5 mM MgCl2, 0.05 mM of each deoxynucleotide triphosphate, 0.32 μM of each primer, 2 U of Taq DNA polymerase (Invitrogen Corporation, Brazil), and 10 μl of clinical specimen. Cycling conditions began with an initial 7 min denaturation step at 94°C, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 45°C for 3 min, and extension at 72°C for 3 min. An additional 7-min extension at 72°C was performed at the end of the 40 cycles. At the first step, 1.4 kbp DNA fragments that is larger than a full length of the ompA gene was amplified. At the second step for nested PCR, 1.2 kbp DNA fragment, a full length of ompA gene was amplified.
Genotyping was performed by Hinf I, Hind III, Cfo I and Hha I restriction analysis of amplified ompA. The precipitated and purified DNA from the remaining 90 μL of PCR product was used for RFLP analysis with restriction enzyme, was prepared in a 20μL reaction mixture containing 3 μg of purified DNA, 15U of enzyme, and 2μL of 10x buffer provided with the enzyme and mili Q water qs, and was incubated overnight at 37°C. After inactivation of the enzyme at 65°C for 10 minutes, the product was electrophoresed through a 3% agarose gel in an electrophoresis apparatus (LKB, Pharmacia, Sweden), stained with ethidium bromide and visualized under an UV transilluminator. (UVP, UK).
Products were electrophoresed through a polyacrylamide gel (acrylamide/bisacrylamide, 29:1; 12 V/cm for 1.5 h) to enable identification of serovars B/Ba, D, E, F, G, K, L1, L2, and C complex (C, J, H, I, and L3). Serotypes C and J were differentiated after digestion with Hinf I. Serotypes H, I, and L3 were separated with Cfo I digestion. Each ompA genotype was defined by testing a reference strain representing each of the 20 serovars. Comparison of the RFLP profiles of each clinical isolate with those of the reference strains was performed to assign a serovar from the genotype of each clinical isolate. Samples unclassified by PCR-RFLP were sequenced and compared with prototype strains. The ompA DNA sequences of unclassified strains were also compared with the prototype strains, B/ TW-5/OT, B/Jali-20/OT, Ba/AP-2/OT, D/ UW-3/CX and D/IC-Cal-8.
Clinical specimen collection was performed by the author’s responsibilities under the agreement based on ethical guidelines on clinical research and the Helsinki Declaration. The study plan was approved by the Research Ethics Committee of International University of Health and Welfare Graduate School.
3. Results
Applied to 15 reference strains, this PCR-RFLP procedure allowed us to determine 13 of the 15 serovars of C. trachomatis. The digested PCR products showed two or three major fragments upon electrophoresis in a 2% agarose gel. Nine patterns were recognized with Hha I, four with Hind III and six with Hinf I. The B and Ba serovars, however, could not be separated by this method. The possibility that clinical isolates frequently have variant patterns was explored by analyzing clinical isolates which were found by culture and antigen detection by EIA to contain Chlamydia species. Thirteen of 15 isolates of nasopharyngeal origin and 11 of 12 isolates of conjunctival origin could be un-equivocally assigned to one of the prototype.
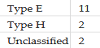
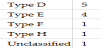
The typing of 15 nasopharyngeal isolates gave the following results: 11 as E, 2 as H, and 2 as unclassified serovars. The typing of 12 conjunctival isolates gave the following results: 5 as D, 4 as E, 1 as F, 1 as H and 1 as unclassified serovar (Table 1 and Table 2). Among the unclassified serovars, two groupings were identified by PCR-RFLP analysis, one was B-like and the rest was D-like. Two unclassified strains showed a type D pattern with Hha I, B, Ba, D, F, G and L1 patterns with Hind III and B, Ba, E, L1 and L2 patterns with Hinf I. One showed B, Ba and E patterns with Hha I, D with Hinf I and the same patterns as the former with Hind III. The B-like strain was identical to each other, the major sequence differences compared to serotype B occurred in VD1.
Comparison of the ompA gene sequences revealed five nucleotide changes, which gave rise to two amino acid alternations in VD1, a single nucleotide change which resulted in an amino acid substitution in VD2 and a single nucleotide change which resulted in no amino acid substitution in VD3. The VD2 and VSD regions of the ompA gene sequence were identical to those in the case of B/Jali-20/OT, which was isolated from a patient with trachoma. In B-like strain, the amino acid sequence of VD1 was identical to that in Ba/AP-2/ OT. Comparison of the ompA gene sequences of the D-like strains with that of D/UW-3/CX revealed two nucleotide changes, resulting in a single amino acid substitution in VS1. The ompA VDs nucleotide sequence was identical to that of D/IC-Cal-8.
4. Discussion
Early diagnosis and appropriate treatment of chlamydial infections may reduce the perinatal complications [11,12]. Sequencing the entire MOMP gene and cataloging the sequences of VDs of all serovars has confirmed the molecular basis of the serotyping procedure and provided a method for determining serovars by PCR-RFLP [1,2]. Genovariants have also been reported for most serovars. The PCRRFLP method for typing allows quick and objective identification of serovars of C. trachomatis [3,13]. C. trachomatis serovars of A B, Ba and C have been predominantly associated with endemic trachoma [1]. In contrast to urogenital chlamydial infection, trachoma is a household disease that has disappeared in many parts of the world because of improved living conditions and hygiene.
The ompA gene PCR- RFLP analysis has some weak points, such as the emergence of atypical restriction patterns due to mixed-genotype infections, artifacts from the enzymatic digestion, and ambiguities due to polymorphisms in the restriction sites or ompA recombinants [10]. The resolution of such atypical patterns requires cumbersome and time-consuming analysis and/or additional runs with different restriction enzymes.
The serovars, D, E, F and H, were similar to those reported in other studies. However, there may be no clear pathogenic distinction between the serovars of endemic trachoma and STI (Figure 3). Unclassified serovars may be new strains or serovariants. There was not enough evidence to be sure that these unclassified clinical isolates were indistinguishable and represented a clone. DNA sequencing or other molecular biological assays seemed to be necessary to analyze unclassified strains. Trachoma strains but not genital isolates carry a deletion or frame shift mutation in a variable region encoding genes for tryptophan synthesis. In subjects infected with serotype E, a T-cell epitopes in VD 3 is recognized significantly less often than in subjects infected with other serotypes of self-limiting follicular conjunctivitis.
Additionally, on the basis of amino acid sequence homology, the classic 15 serovars of C. trachomatis have been classified into another 3 genetic groups: group B (B, D, E, L1 and L2), group C (A, C, H, I, J, K, L1 and L3) and an intermediate group (F and G). Serovars D-K are commonly associated with urogenital infections and perinatal conjunctivitis or pneumonia worldwide [11]. In women a broad spectrum of clinical manifestations can be present due to urogenital C. trachomatis infections including urethritis and cervicitis and if left untreated, the ascending infection may cause secondary complications in the upper genital tract, including pelvic inflammatory disease and infertility [9,14]. Serovars L1-L3 are associated with LGV, a systemic disease most prevalent in tropical and subtropical areas. Nevertheless, several outbreaks among homosexual male populations in western countries have been reported recently, indicating the merging role of LGV [15,16].
Currently C. trachomatis has evolved to include 19 serovars based on antibody typing of MOMP with over 60 ompA genotypes [7], the gold standard typing technique for all Chlamydiaceae spp. Ocular infections include trachoma, a chronic ocular disease, and neonatal inclusion conjunctivitis, an infection acquired during passage through a C. trachomatis- infected birth canal. Urogenital strains cause not only ocular infections, which usually present as unilateral conjunctivitis, but also can ascend from the endocervix to cause sequelae such as pelvic inflammatory disease, infertility and ectopic pregnancy. Rectal infections can progress to proctitis and inguinal syndrome [17]. While the later is caused primarily by the LGV strains L1-3, L2a, L2b, and L2c, the former can be caused by most C. trachomatis strains, although strains B, Ba, and C are usually not detected in the urethra, endocervix or rectum [18-20]. Strain A is the only strain that is confined to the ocular mucosa [9] .
Although ompA genotyping can be informative, the gene represents a mere 0.1% of the genome and is subject to immune selective pressure and recombination [10,21,22]. Partial and whole genome sequencing (WGS) have added considerably to our knowledge of the diversity of C. trachomatis and evidence for recombination [23]. Patients infected with C. trachomatis have significant risk of being infected with U. urealyticum and HIV, suggesting screening of C. trachomatis along with other STIs. Further, genotyping studies are required to understand the geographical distribution and introduction of new serovar in the community [20,24-26].
5. Conclusions
The sequences of MOMP gene for all 15 classic serovars allowed the construction of restriction endonuclease cleavage-site maps that confirm the fragment-size patterns observed by electrophoresis. Sequencing the entire MOMP gene and cataloging the sequences of VDs of all serovars has confirmed the molecular basis of the serotyping procedure and provided a method for determining serovars by PCR-RFLP. Although geno-variants have also been reported for most serovars the PCR-RFLP method for typing allows quick and objective identification of serovars of C. trachomatis. There maybe no clear ocular pathogenic distinction between the serovars of endemic trachoma and those associated with STI. Antigenic variations of C. trachomatis were also considered among the strains from nasopharyngeal and conjunctival origins.
Competing Interests
The author declare that there is no competing interests regarding the publication of this article.
Acknowledgments
The author express the gratitude to Masami Ikehata and Hideomi Asanuma at the Department of Pediatrics, Sapporo Medical University School of Medicine, Japan, for their valuable technical assistance and comments.