1. Introduction
Blood circulation infections are amongst the major causes of morbidity and mortality in humans. Blood culture is a golden standard life-saving method for early diagnosis and appropriate antimicrobial treatment in critically ill patients, especially in the intensive-care-unit patients and in the patients with neutropenia. Various systems have been developed for blood culture testing. But there are a limited number of systems routinely used worldwide with high performance that are capable of detecting the microorganism automatically [1-3] . These systems detect the presence of bacteria measuring the amount of carbon dioxide present in the bottles with detecting (i) pressure, (ii) colorimetric, and (iii) spectrophotometric changes [3]. Commercial systems are widely used in the world, such as BacT/Alert® 3D (BioMérieux, France) and Bactec 9240™ System (BD Microbiology, Cockeysville, MD). The VersaTREK® System (Trek Diagnostic Systems, Cleveland, Ohio) system is less preferred [3-5] . Each laboratory prefers its own system depending on various reasons, such as price, capacity, ease of use and sensitivity for bacteria detection.
Signaling of the blood culture system even if there are no microorganisms in the culture bottle is defined as “false positivity”. Various factors related to the patient or the microorganisms can cause false positivity [3-5] . All false positive repetitive alarms can cause misinterpretation errors, inappropriate antibiotic use, additional examination, prolonged hospital stay, and increased treatment costs [3]. In the present study, it was aimed to identify the factors related to the patient or to the microorganisms which may cause false positivity in the automated blood culture systems, and to evaluate the parameters to be used in interpreting the results correctly.
2. Materials and Methods
The study was carried out in the Gaziosmanpaşa Hospital Microbiology Laboratory of Istanbul, Yeni Yüzyil University Faculty of Medicine between February 2016 and February 2017. In this study, "Fully Automated Bact/Alert 3D" (BioMérieux, France) system and culture bottles compatible with this system were used as the blood culture method. In the laboratory, technical staff have been trained periodically on blood culture technique, and the blood cultures have been collected and processed following the recommendations of Centers for Disease Control and Prevention (CDC) [1,6].
2.1 Processing of the blood cultures
Appropriate amount of blood culture samples were taken into the bottles (8-10 cc for adults, 1-3 cc for children), and data of the patients were written on the bottles. They were transferred to the microbiology laboratory immediately. The blood count tests were performed within the first 24 hours. Blood cell counting were evaluated using fluorescence flow cytometry (FFC) method (Sysmex XT-1800i and XT-2000i, Japan).
When the positive alarm signal was received from the device, the presence of the microorganism was first investigated using the Gram staining method. In spite of the presence of a growth signal, the absence of microorganisms in the gram preparations and the absence of growth in the incubation plates were accepted as false positivity [3]. If the microorganism was not seen in the Gram staining, the prepared preparations were stained using acridine orange and re-evaluated for the presence of microorganisms.
When a positive signal indicating that a culture was positive, it was inoculated onto a 5% sheep blood agar and an EMB agar media (Salubris, Turkey), and the plates were incubated for five days. In addition, a couple of preparations were prepared on glass slides for stained microscopic examination. When the growth signal was positive but the microscopic examination didn’t show any microorganisms, the cultures were additionally inoculated onto a chocolate agar and a Sabouraud Dextrose Agar (Salubris, Turkey) media, and the incubation was extended to 14 days. In case of the absence of microorganisms in the Gram and acridine orange-stained preparations and in the absence of growth in the media at the end of the incubation period, the signal from the automated system was assessed as false positivity [2,4,5]. Repetitive blood cultures of the same patient were excluded from the study.
2.2 Data analysis and ethical permission
The study group consisted of the patients who do not have any microorganisms in the Gram-stained preparation despite the growth signal in the culture system. Patients who were sampled in which the growth signal was confirmed consisted the control group as true positive cases, and this control group was selected by random sampling method. Mann Whitney U test and Chi-Square test were used for statistical analysis. Permission was taken from Istanbul Yeniyüzyil University Ethics Committee with the number 10.03.20707 for the study.
3. Results
A total of 9216 patients with aerobic blood culture tests were included in the study, and 1839 (19.9%) of those gave a positive growth signal. False positivity was detected in 69 (0.75%) of the blood culture bottles that indicated a positive growth signal. The mean incubation time for positive growth signal was 20 hours (with an interval between 3-87 hours) in the bottles giving a true positive growth signal. The mean incubation time for positive growth signal was 3 hours (3-66) in the bottles giving false positive growth signal, and the difference was statistically significant (p<0.001) (Table 1).
A total of 88.9% of blood cultures that provided a true positive growth signal indicated between 6-10 hours of the incubation. Only one blood culture bottle with a true positive signal indicated within the first six hours. In contrast, 90.5% of the blood cultures that gave false positive growth signals signaled within the first four hours of the incubation.
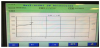
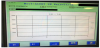
At the moment of the first signals were taken from placed bottles in the device, the true positive bottles had a tendency of increase in the growth graphs (Figure 1). The negative bottles’ graph indicated that there were no microorganisms in the bottle, and tended to be horizontal (Figure 2). The false positive growth signals tended to be a highly rising graphic within the short term graphs (Figure 3).
When the distribution was examined according to the services, it was observed that the highest number of the samples with false growth signal was taken from intensive care units and hematology services.
4. Discussion
False positivity of the cultures cause delays in accurate and rapid diagnosis of blood circulation infections. Thus, appropriate antimicrobial therapy cannot be made, and the possible mortality rates as well as treatment costs may increase [3,5]. When these systems detect the presence of a bacterium, it is also important to assess the results with different parameters to be sure whether the detected microorganism is an infection factor in the patient.
In our study, we found the false positivity rate of blood culture to be low as 0.75%. Amongst the studies that used the Bact/Alert blood culture system, Mirrett et al. [7] found the false positivity rate as 6%, Karahan et al. [8] found as 1%, and Mehli et al. [9] found as 0.63%, while Drayer et al. [10] did not report any false positivity for this system. Amongst the studies that used the Bactec Blood Culture System, Qian et al. [11] reported the false positivity rate as 1.6%, Kurtoğlu et al. [12] reported as 0.2%, Nolte et al. [13] as 2.2%, and Ziegler et al. [14] as 1.4%. However, Smith et al. [15] reported a slightly higher false positivity than the other systems in the study that they evaluated both systems, as 0.5% for the Bactec System, and as 1.2% the Bact/Alert. In the studies evaluating the Versa TREK, Dreyer et al. [10] reported false positivity rate as 7.9%, and Mirrett et al. [7] reported as 1.6%. When the results of these studies are evaluated, it is seen that the system with the highest false positivity rate is Versa TREK [7,10]. Despite the various reports, it can be said that the false positivity rate in the Bactec System is generally higher than Bac/Alert (Bactec: 0.2-2.2% average 1.18 vs Bact/Alert: 0-1.6% on average 0.88) [7-15] . Except for the study conducted by Smith et al., we found that Bactec's false positivity rate was slightly higher than Bact/Alert in blood culture systems based on CO2 measurements. When the Versa TREK system based on pressure measurement is considered, it is understood that higher false positivity rates are reported than these two systems mentioned above [15].
It has been reported that leukocytes produce carbon dioxide as a part of their metabolic processes, causing false positive growth signals in the automated systems that spectrophotometrically detect the increase in the amount of carbon dioxide, such as BacT/ Alert 3D [5,11,13, 15-17]. In our study, the numbers of leukocytes, neutrophils, and immature granulocyte were found to be statistically higher amongst the factors due to the patients which may cause false positivity. In addition, a group of the patients with normal number of leukocytes and false positive blood culture was the intensive-care-unit patients who were taking antibiotics, and that might be another factor of false positivity [15].
Among the causes of false positive growth in blood culture systems due to the microorganisms include; (i) slow and fastidious microorganisms, (ii) microorganisms that cannot be cultured, (iii) anaerobic microorganisms, (iv) bacteria more sensitive to environmental conditions such as S.pneumoniae, (v) culture bottles contaminated with microorganism DNAs, and (vi) bacteria transforming into L-form due to antibiotic use [2,3]. In order to identify a small number of microorganisms, extension of the incubation period for factors belonging to microorganisms, cultivation of enriched media to facilitate production, molecular or acridine-staining methods are recommended [15,17]. As a mistake of the bacteriology laboratory, the presence of bacteria requiring additional processing for growth in routine bacterial cultures is one of the factors considering the false positivity. For instance, a self-autolytic microorganism such as S. pneumoniae may also cause this mistake [2-4,12]. However in the present study, Gram and acridine orange stains were used to exclude this possibility, and no microorganisms were observed with microscopic examination. In addition, chocolate agar was used to cultivate by enriching and SDA was used for fungi, and growth controls were followed up for a period of two weeks inspite of the recommendation of five days. However, no bacteria were detected with all these methods. In this study, no culture was made for anaerobic microorganism, no anaerobic agents were considered because no bacteria were observed in staining methods. When false positive growth signal is detected in the automated blood cultures system, searching for DNA by molecular methods are also suggested [5,7,15,18].
In this study, molecular methods could not be used due to the reasons such as lack of technically experienced staff, time consuming and cost. Nonetheless, in order to show the presence of nucleic acids belonging to microorganisms, acridine orange staining method was used. The acridine orange stain can be linked to the nucleic acids of the bacteria, a small number of microorganisms can be detected. Acridine orange is more sensitive to uncover microorganism than the Gram stain [7,8,19]. Acridine orange staining method is particularly useful in the identification of members of the genus Campylobacter, Brucella spp. and Mycoplasma [19]. In our study, no microorganisms were found in negative bottles despite staining with acridine orange. Although the incubation period of blood culture bottles was extended for two weeks, which were thought to give false positive growth signal, there was no growth at the end of the incubation period. Thus, slow-biogenic fastidious microorganisms such as Brucella spp. were not overlooked [1-3,12, 20]. Microorganisms, which are common in the automated blood cultures, give true positive growth signals within the first 24-45 hours of incubation and show growth on the inoculated plates [2,8]. In our study, the mean growth time in blood cultures which revealed true positive growth signals was 20 hours (3- 87 hours). A total of 88.9% of blood cultures signaled after 10 hours of incubation. Only one patient's blood culture bottle had a real positive signal within the first six hours. In the majority of false positive blood cultures, the growth signal is detected within the first four hours. False positive blood cultures are rarely seen after 48 hours [3,5,8]. In our study, we found that 90.5% of false positive blood culture bottles signaled within the first four hours (3-66 hours) with a statistically significance. This situation suggests that if there is a positive signal within the first four hours, it may be a false positive result.
In this study, 19.9% of the blood culture bottles provided a true positive growth signal. This rate is compatible with other studies in this subject, although it varies according to the characteristics of the patients in different regions and in different hospitals [7,9,15].
As a result; in order to interpret the blood culture results correctly, it should be considered as a false positive growth signal when the blood culture signals within the first four hours, and/or in cases of high numbers of leukocytes, neutrophils or immature granulocytes. If the clinical microbiology laboratory detects a positive signal in a blood culture within the first four hours, the laboratory should revise the blood cell counts of the patient in terms of false signal positivity. Thus, we think that unnecessary examinations can be prevented, and the length of hospital stay, treatment costs, and unnecessary antibiotic use can be reduced as well.
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
DBT: Proposed and designed the study,
TK: Collected, analysed
and interpreted the data,
DG, FK, KŞ: Participated in proofreading of
the manuscript and in critical revision.
All authors read and approved
the final manuscript.