1. Introduction
Enterobacteriaceae are among the most important etiological factors of nosocomial and community acquired infections. The most commonly isolated pathogens are Klebsiellapneumoniae, Escherichia coli and Enterobacter spp. Salmonella and Shigella spp [1]. The prevalence of Gram-negative microorganisms that are resistant to different antibiotics and the resultant deficit in antibiotics that emphasizes the urgent need for novel therapeutic agents for the treatment of Gram-negative infectious agents [2]. Beta-lactams and fluoroquinolones constitute the main therapeutic agent of choices to treat infections caused by these pathogens. However, the development of resistance to these compounds, especially, by Gram negative microorganisms constitutes a noticeable success of bacterial survival [3,4,5]. Quinolones resistance complicates the treatment of infections caused by ESBL-producing Enterobacteriaceae strains [6]. The most important mechanisms involved in quinolones resistance in Enterobacteriaceae are (i) accumulation of chromosomal mutations primarily in the quinolone resistance-determining regions (QRDRs) of the target genes, gyrA which encodes DNA and parC which encodes topoisomerase IV [7]. (ii) Up regulation of the native efflux pumps which decreases the intracellular drug accumulation [8] alone or in addition to decreased expression of outer membrane proteins [9]. Three plasmid-mediated fluoroquinolone resistance (PMQR) mechanisms have been described; including (i) the Qnr (qnrA, qnrB, qnrS, qnrC and qnrD proteins, (ii) the aac(6')-Ib-cr enzyme, and, (iii) QepA and oqxAB plasmid-mediated efflux pumps [5]. ESBLproducing isolates are commonly associated with PMQR genes in Enterobacteriaceae [6]. Use of efflux pump inhibitors is a unique anti-resistance approach that can return activity for different families of antibiotics. However, their clinical use is difficult due to the toxicity, stability, selectivity and bioavailability of available molecules [10]. Phenylalanine-arginine β-naphthylamide (PAβN) is a well-studied efflux pump inhibitor (EPI) that is routinely combined with fluoroquinolone antibiotics. As a result of the competition between PβNA and the antibiotic, PβNA is extruded outside by the efflux pump and the antibiotic reaches its effective concentration inside the cell [11]. The aim of this study was to investigate the most common mechanisms of quinolones resistance in ESBL-producing Enterobacteriaceae clinical isolates using both phenotypic method by testing the effect of PβNA (EPI) and genotypic methods by using multiplex- PCR to detect plasmid mediated (PMQR) and PCR-RFLP assay to detect mutations in the quinolone-resistance determining regions of gyrA and parC. The detection of mutations associated with reduced susceptibility to fluoroquinolones by PCR-RFLP is well thought-out in our country as it serves as a specific, rapid, inexpensive, and simple testing alternative to sequencing assays.
2. Material and Methods
2.1 Ethics statement
The present study was approved by the Faculty of Medicine Cairo University Hospital, Egypt. Written informed consent was not necessary for this retrospective study, as it was part of our standard microbiological routine. Patient data were anonymous for the purposes of this analysis, and all confidential patient information was protected in accordance with Egyptian law.
2.2 Hospital setting
The study was conducted at Cairo University Hospital, which serves patients in Cairo (Egypt) and provides medical and surgical care in all medical specialties. The study took place from October 2012 to September 2013. The study conforms to the relevant regulatory standards and is in accordance with the recommendations of the Clinical and Laboratory Standards Institute (CLSI) guidelines.
2.3 Clinical isolates (selection and identification)
Out of 1766 clinical bacterial isolates collected from October 2012 to September 2013, a total of 219 Enterobacteriaceae clinical isolates were ESBL- producers, nalidixic acid and ciprofloxacin resistant. All isolates were collected from inpatients admitted at Kasr Al-Ainy Cairo University Hospital, Egypt. All 219 isolates were processed and identified by the standard procedures of the Clinical and Laboratory Standards Institute guidelines [12] and were identified to the species level using MALDI-TOF/MS with score values > (1.9) using the Bruker software microflex RTC version 3.1 and then stored in brain heart infusion broth with 15% glycerol at -60ºC until further analysis.
2.4 Phenotypic detection of Quinolones resistance
All 219 Enterobacteriaceae isolates were subjected to phenotypic detection of quinolones resistance.
2.5 Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by the disc diffusion method (Modified Kirby-Bauer technique) using Muller Hinton agar, aerobic incubation at 35oC for 16– 18 h. Antimicrobial discs, Imipenem (10 mcg), Meropenem (10 mcg), Gentamicin (10 mcg), Ciprofloxacin (5 mcg), Amikacin (30 mcg), Cotrimoxazole (25 mcg), Cefepime (30mcg), Cefotaxime (30mcg), Cefotaxime+ Clavulanic acid (30/10mcg), Aztreonam(30mcg), Ceftazidime (30 mcg), Ceftazidime + Clavulanic acid(30/10 mcg), Amoxacillin- Clavulanic acid (20/10mcg), and Cefoxitin (30 mcg) were obtained from Oxoid Co. (Oxoid Limited, Basingstoke, Hampshire, England). Phenotypic screening and confirmation of Extended-Spectrum β-LactamaseExtended-Spectrum β-LactamaseESBL production was done using the combined disc tests [12].
2.6 Minimal Inhibitory Concentration
Minimal inhibitory concentration of nalidixic acid (NA) and ciprofloxacin (CIP) was determined by broth microdilution method following CLSI guidelines 2012. CIP and NA MICs were measured in microdilution plates with a final volume of 200 μl/ well. 100 μl Mueller-Hinton broth was added in each well plus an extra 74.4 μl only in the first well and 25.6 μl ciprofloxacin from a 2 mg/ml stock solution. Serial dilutions were done and 100 μl of bacterial inoculum with 105 CFU/ ml was added in each well, obtaining ciprofloxacin concentration of 128 μg/ ml in the first well and 0.06 μg /ml in the last one. In case of Nalidixic acid 10.24 μl was added in the first well from a 5 mg/ml stock solution. After serial dilutions and addition of bacterial inoculum with 105 CFU/ml in each well; Nalidixic acid concentrations of 128 μg/ ml in the first and 0.06 μg/ ml in the last wells were obtained. The lowest antibiotic concentration that inhibits the growth was considered as the MIC value.
2.7 Phenylalanine-arginine β-naphthylamide (PaβN) efflux pump inhibitor
According to the CLSI (2010) recommendations, before and after addition of PβNA (Sigma-Aldrich Co, St Louis, MO, USA) the lowest antibiotic concentration that inhibits the growth was considered as the MIC value. One hundred μL of Mueller-Hinton broth containing the bacterial suspensions was added to the wells of a sterile microdilution plate. By adding 50 μL of suitable concentrations of CIP and NA to the first line of wells, serial dilutions were performed. 20μL constant PβNA concentration solution was added (20 μg/ml) to each well. At least four fold decreases in the MIC values of ciprofloxacin and nalidixic acid was evaluated as the presence of the efflux pump [13].
2.8 Genotypic detection of Quinolones resistance
Out of a total of 219 isolates, 28 isolates showed MIC changes after addition of PβNA, 5 isolates were added as a negative control. A total of 33 isolates were subjected to PCR assay of the PMQR genes, as well as PCR amplification of the QRDRs of gyrA and parC, followed by RFLP to detect mutations in gyrA and parC.
2.9 PCR amplification
PCR amplifications of the quinolone resistance determining regions (QRDRs) of gyrA and parC were carried out using the primers listed in Table 1. The primer used to amplify a 344-bp fragment containing the QRDR of the gyrA gene. The conditions were 94°C for 60 sec, 55°C for 45 sec, and 72°C for 60 sec for 30 cycles. For parC gene, amplification was carried out for 35 cycles at 94 °C for 30 sec, 55 °C for 30 sec, and 72 °C for 30 sec to amplify a 168 bp fragment [14]. PCR products were submitted to electrophoresis on a1.5% agarose gel containing ethidium bromide to visualize the amplified bands under UV.
2.10 Restriction Fragment Length Polymorphism (RFLP)
All PCR products positive for gyrA and parC genes were further analyzed by digestion with Hinf I (Thermo scientific FastDigest restriction enzyme, Fermentas-Lithuania) to identify mutations [15]. The wild-type gyrA contains the artificial Hinf I cleavage site. Consequently, Hinf I digests the amplified 344-bp products to produce two fragments for gyrA 210 and 272 bp respectively, and also for the wild type parC HinfI digest 168 PCR product for 32 and 65bp respectively. Ten μL of each amplification product was mixed with 1 μL of Hinf I, 2 μL of 10 X fast Digest green buffers and 17 μL of water and incubated at 37 C for 5 min. Ten micro-liters of the digested fragments were run in 3% agarose gel. The gel was stained with ethidium bromide and the DNA bands were visualized with UV transilluminator. DNA ladder (50 pb and 25 bp) (Promega1) was used as a molecular weight marker.
2.11 Detection of Plasmid Mediated Quinolones Resistance Determinants (PMQR)
Screening of the five PMQR determinants was carried out by two sets of multiplex PCR amplification, one for qnrA, qnrB, and qnrS and the other for aac (6’)-Ib and qepA [14].Also Detection of oqxAB was performed as described elsewhere[16].
2.12 Statistical analysis
The resulting data was analyzed using SSPS version 13 program. Nominal data was expressed as frequency and percentage and compared using Chi square test. The Pvalue < 0.01 was considered significant.
3. Results
Out of 1766 clinical Enterobacteriaceae isolates, 219 (12.4%) isolates were ESBL producers and quinolone resistant, including; Escherichia coli 98 (44.7%), Klebsiella pneumoniae 92 (42%), Salmonella enterica 8 isolates plus one isolate Salmonella enteritidis, one isolate Salmonella choleraesuis and one isolate Salmonella bongori total 11 (5%), one isolate Enterobacter ludwigii plus two isolates Enterobacter. cloacae total 3 (1.4%), Proteus mirabilis 8 (3.65%) and Shigella flexneri 2 (0.9%). The most common site of isolation was the urine specimens 112 (51.1%) (The P-value was significant P < 0.01), wound specimens 65 (29.6%), sputum specimens 16 (7.3%) followed by stool specimens 13 (5.9%). The distribution of clinical isolates according to the clinical specimens are shown in Table 2. The presence of co-resistance among different classes of antibiotic families was significant (The P-value is 0.000346) except for carbapenems and polymyxin classes. In our study minimum inhibitory concentrations (MICs) of ciprofloxacin antibiotic and nalidixic acid before and after addition of efflux pump inhibitor PaβN resulted in reduction of MIC in total 28 isolates. In E. coli isolates adding PaβN resulted in reduction in MIC by more than 2 log2 dilution in 9 samples for ciprofloxacin antibiotic and in 4 samples for nalidixic acid (The P-value is 0.145483, the result is not significant at P < 0.10). While addition of PaβN to Klebsiella pneumoniae isolates resulted in reduction of MIC by more than 2 log2 dilution in 8 isolates for ciprofloxacin antibiotic and in 5 isolates for nalidixic acid (P-Value is 0.997521, the result is not significant at P < 0.10). Moreover addition of PaβN to the Enterobacter spp isolates resulted in reduction of MIC by more than 2 log2 dilution in 2 samples for both ciprofloxacin and nalidixic acid antibiotic (P-value is 1, the result was not significant at P < 0.10). Whereas addition of PaβN to the Proteus mirabilis isolates resulted in a reduction of MIC by more than 2 log2 dilution in 2 samples for ciprofloxacin and in 2 samples for nalidixic acid antibiotic, the result is also insignificant at P < 0.10. Also addition of PaβN to the Salmonella spp isolates resulted in reduction of MIC by more than 2 log2 dilution in 6 samples for both ciprofloxacin and nalidixic acid antibiotic by a percent 100% ( The result is not significant at P < 0.10). Finally addition of PaβN to the Shigella flexneri isolates resulted in reduction of MIC by more than 2 log2 dilution in the 1 samples for both ciprofloxacin and nalidixic acid antibiotic by a percent 100% (The P-value is 0.252656, The result is not significant at P < 0.10). Thirty three isolates (28 of them showed MIC changes after addition of PβNA in addition to 5 isolates added as a negative control) were selected for further PCR work up, including PMQR genes, as well as PCR amplification of the QRDRs of gyrA and parC, followed by RFLP to detect mutations in gyrA and parC. By PCR-RFLP assay gyrA gene mutation was detected in 24/33 (72.7%), while par C gene mutation was detected in 3/33 (9.1%).
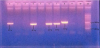
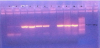
The prevalence of 85.7% of PMQR genes and qnr-genes were identified in 13/33 (39.3%) of the clinical isolates as shown in (Figure1), one isolate contained qnrA gene only, one isolate contained the qnrB gene only, one isolate contained both qnrA and qnrS genes and two isolates contained both qnrB and qnrS genes. Aac(6')-Ib gene was detected in 72.7 % (24/33) of the clinical isolates as shown in (Figure 2). The prevalence of aac(6')-Ib-cr was 72.7% (8/11) among E. coli, 88.8%(8/9) among Klebsiella pneumoniae, 66.6% (2/3) among Enterobacter spp., 66.6% (2/3) among Proteus mirabilis and 66.6% (4/6) among salmonella spp clinical isolates. oqxA gene was detected in 51.5 % (16/33) of the clinical isolates. The prevalence was 18.1% (2/11) among E. coli, 66.6% (2/3) among Enterobacter spp., 33.3% (1/3) among Proteus mirabilis and 50% (3/6) among Salmonella spp. Klebsiella pneumoniae isolates showed positive results for oqxA gene by 100% (8/9). Whereas oqxB gene was detected in 21.2 % (7/33) of the clinical isolates. QepA gene was detected in only one Klebsiella pneumoniae isolate which was highly resistant to both antibiotics before and after the addition of PaβN as shown in (Figure 3).
All E. coli Enterobacter, Proteus and Salmonella isolates showed negative results for the OqxB gene. Isolates of positive PCR results for efflux pump genes oqxAB showed MIC changes after addition of efflux pump inhibitor PaβN in 88.2% (15/17) for ciprofloxacin and 70.5% (12/17) for nalidixic acid. The results of multiplex-PCR and PCR-RFLP for the tested Enterobacteriaceae clinical isolates are presented in Table 3.
* isolates which selected for PCR screening.
4. Discussion
The occurrence of antibiotic-resistant bacteria of all genera has been on the rise. Efflux pumps are transport proteins involved in the extrusion of toxic substrates (including virtually all classes of clinically relevant antibiotics) from within cells into the external environment. The emergence and spread of MDR Enterobacteriaceae that cause human infections is increasing worldwide. The most common problematic pathogens in this family are: Klebsiella pneumoniae, Escherichia coli and Enterobacter spp. They are identified mostly in hospital settings but also as a source of community-acquired infections [1]. Urinary tract infections (UTI) are among the most common bacterial infections in various parts of the world with high medical costs. In this study the most common site of isolation was the urine 112 (51.1%) [17].
In this study, the MICs of ciprofloxacin antibiotic and nalidixic acid antibiotics before and after addition of efflux pump inhibitor PaβN were measured in 219 strains with fixed concentration of PβNA (20 ug/ml). Changes were detected in only 28 isolates. Helaly et al. [18] found that by increasing the concentration of PAβN, there is progressive reduction in MIC of ciprofloxacin and levofloxacin which is the same for concentration 25 and 50 μg/ml (8-and 16- fold for ciprofloxacin and levofloxacin, respectively), while at concentration of 100 μg/ml a more reduction in the MIC is noticed reaching 16-fold for ciprofloxacin and 32-fold for levofloxacin (64-fold for E30). While Lunn et al. [19] reported that when the MICs of ciprofloxacin were measured in the presence of PAβN, ten of thenalidixic acid resistant isolates showed a 1.3- to 2-fold decrease, whereas no change was detected in any of the other seven isolates. In all but two isolates (MIC of 0.094 μg/ml), susceptibilities to ciprofloxacin remained decreased in the presence of PAβN. Glatz [20] observed that at least four-fold reduction in ciprofloxacin MICs was found in the presence of PAβN in 79% of representative isolates; at least eight fold reductions in ciprofloxacin MICs in the presence of PAβN (PAβN+) was detected in 37% of representative isolates. In another study conducted by Lavigne et al. [21] on five patients presented imipenem susceptible E. aerogenes strains, then intermediate or resistant isolates. They observed that the PAβN addition reduced the MICs for Ofloxacin in all strains. Another study carried out by Yedekci et al. [13] noted changes in nalidixic acid and ciprofloxacin MIC values in the presence of fixed concentration of PAβN (20 pLg/ml).
All the 33 strains were subjected to PCR-RFLP with the same PCR primers and restriction enzymes. PCR amplification of gyrA and parC was successful for all 33 clinical strains including the positive controls which generated products with the expected amplicon sizes of 344 and 168 bp. Our PCR-RFLP method is simple because the PCR primers and restriction enzymes are identical and the method is compatible with all 6 Enterobacteriaceae species. Therefore, the level of quinolones resistance for all 6 Enterobacteriaceae species may be determined without previous species identification from clinical specimens. The PCR-RFLP method provided results within 5 h. Since plasmid-mediated quinolone resistance (PMQR) was first described in 1998, four types of PMQR determinants have been identified: qnr, aac(6')-Ib-cr, qepA, and oqxAB [22].The qnr genes are transferable genes that confer low-level quinolone resistance by protection of topoisomerase, qnrA had an additive effect of a 10-fold increase in the minimum inhibitory concentration (MIC) whatever the number of topoisomerase mutations, and qnrS was additive to qnrA with a further 2- to 10-fold increase in the MIC [23].
In Egypt a study carried out by Hassan et al. [24] who found that out of 30 ESBL producers E. coli isolates, 8 (26.6%) were positive for qnr genes, and the qnrA1-, qnrB1 and qnrS1-type genes were detected alone or in combination in 5 (16.6%), 7 (23.3%) and 5 (16.6%) isolates, respectively. Recently, the association of aac(6')-Ib-cr with genes encoding the beta-lactamase CTX-M-15 or other ESBLs has been reported [25]. The qepA gene, together with the qnr family and aac(6')-Ibcr, is the third recently detected plasmid-borne determinant of resistance to the fluoroquinolones. These genes confer only lowlevel resistance, but their presence could potentially facilitate evolution of the bacterial host toward higher levels of resistance by mutational alterations in the target type II topoisomerases [26]. qepA gene was detected in only one Klebsiella isolate in this study which was highly resistant to both antibiotics before and after the addition of the efflux pump inhibitor PaβN. oqxA/OqxB is highly prevalent in diverse MDR K. pneumoniae of human origin. This efflux pump may be an important factor contributing to the MDR profile of K. pneumoniae, and to its versatility as a zoonotic and nosocomial colonizer [27]. Among the 83 K. pneumoniae strains, Taherpour and Hashemi [28]. found that 48 (57.5%) were ESBL positive. The prevalence of both oqxA and oqxB detected in K. pneumonia was high: 50 (60.2%) and 50 (60.2%), respectively. In this study, fosfomycin and tigecycline were more active than other antibiotics Isolates positive for both oqxA and oqxB were regarded as oqxAB positive as the oqxAB is encoded by oqxA and oqxB in the same operon [22].
5. Conclusion
The presence of co-resistance among different classes of antibiotic families was significant among the ESBL-producing Enterobacteriaceae. The PaβN was an effective phenotypic screening method for quinolones resistance. The data obtained suggests the wide occurrence of PMQR genes in clinical ESBL –producing ciprofloxacin resistant Enterobacteriacae isolates. Also the present study provides sufficient data suggesting that PCR-RFLP methodology is a simple and rapid method (it can be performed within 5 hours) for the detection of ciprofloxacin-resistant strains useful for clinical diagnosis and epidemiological studies. Routine surveillance of microbial population to determine the extent of antibiotic resistance should be conducted to provide suitable treatment guidelines.
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
All authors contributed to, designed, and coordinated the study.
Study concept and design: MK, DI, and SM. Microbiological analysis
of the samples: RE, DI, SE and SJ.
Acquisition of data: RE, DI, and SE. Analysis and interpretation of
data: SJ, RE, DJ, SE, and SM.
Drafting of the manuscript: GK, RE, DI, and SE. Critical revision of
the manuscript for important intellectual content, RE, DI, SE, and GK.
Study supervision: MK, RE, DI, SA, SJ, and SE. All authors read and
approved the final manuscript.
Acknowledgments
The authors acknowledge the Faculty of Medicine, Cairo University for technical and general support.